Memory Profiling raw ColBERT and RAGatouille
memory-profiler library to log memory using for different indexing functions for raw ColBERT and RAGatouille indexing operations for 100k, 250k, 500k, 1M and 2M collection sizes. In general, RAGatouille uses more memory than raw ColBERT.
Background
A disclaimer: this is the first time I’ve done memory profiling, and while I’ve probably spent 8-10 hours poring through the RAGatouille and ColBERT codebases I still consider myself a beginner, and don’t have a solid mental model of how indexing (and search) work.
With that out of the way, let’s dig in!
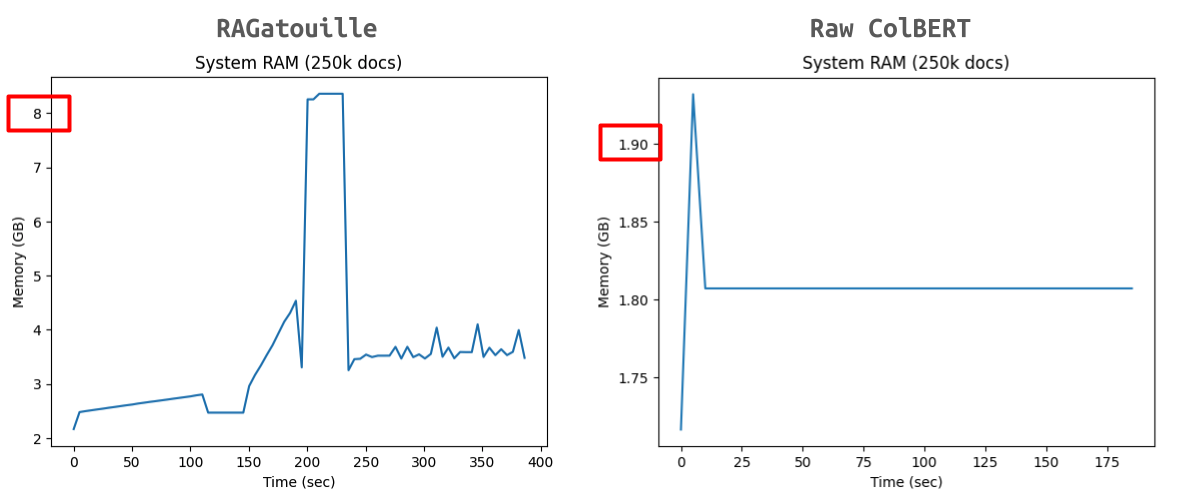
In a previous blog post I used psutil.Process().memory_info().rss in a separate thread to monitor memory usage while indexing 100k, 250k, 500k, 1M and 2M documents from the Genomics datasets (via UKPLab/DAPR) with RAGatouille. I have also run this for raw ColBERT. Here’s an example comparison (for 250k docs on an RTX6000Ada instance) with RAGatouille on the left and raw ColBERT on the right:
While the peak memory increased with number of documents, they all follow the same trend. ColBERT always has a significantly lower peak memory. The ColBERT runs in total took about an hour and the RAGatouille runs took about 1.5 hours. Comparison of all collection sizes can be seen in this folder.
In this blog post I go deeper and use the memory_profiler package to understand how much memory is being consumed by different functions down the chain of calls when you index 100k, 250k, 500k, 1M and 2M documents using raw ColBERT and RAGatouille. For all of these runs I use a RTX6000Ada instance on Jarvis Labs. When using RAGatouille, I execute all runs with use_faiss=False (since that’s the default value in RAGatouille) and runs of 100k, 250k and 500k with use_faiss=True.
Repo Setup and Installation
Since I needed to add the @profile decorator above each function I wanted to profile, I created my own forks of the raw ColBERT and RAGatouille repos and created a profiling branch. Since RAGatouille is built on top of ColBERT, I switched the colbert-ai dependency in my RAGatouille fork from "colbert-ai>=0.2.19" to:
"colbert-ai @ git+https://github.com/vishalbakshi/ColBERT.git@profiling"I also added memory-profiler as a dependency for both ColBERT and RAGatouille.
I used the terminal for all experiments. Here are the commands to install RAGatouille:
python -m venv ragatouille-env
source ragatouille-env/bin/activate
git clone -b profiling https://github.com/vishalbakshi/RAGatouille.git
cd RAGatouille
pip install -e .
pip install datasets
pip uninstall --y faiss-cpu
pip install faiss-gpu-cu12Note that I uninstalled faiss-cpu and installed faiss-gpu-cu12.
Here are the commands to install ColBERT (which took considerably more effort, and assistance from Claude, to figure out):
git clone -b profiling https://github.com/vishalbakshi/ColBERT.git
cd ColBERT
conda env create -f conda_env.yml
conda init
source ~/.bashrc
conda activate colbert
pip install -e .
conda remove -y --force pytorch torchvision torchaudio cudatoolkit
pip3 install torch torchvision torchaudio --index-url https://download.pytorch.org/whl/cu118
apt-get update
apt-get install -y gcc-11 g++-11
export CC=gcc-11
export CXX=g++-11I had to uninstall pytorch, torchvision, torchaudio, cudatoolkit and reinstall them to resolve the following error:
File "/home/ColBERT/colbert/utils/utils.py", line 3, in <module>
import torch
File "/root/miniconda3/envs/colbert/lib/python3.8/site-packages/torch/__init__.py", line 218, in <module>
from torch._C import * # noqa: F403
ImportError: /root/miniconda3/envs/colbert/lib/python3.8/site-packages/torch/lib/libtorch_cpu.so: undefined symbol: iJIT_NotifyEventThe last four commands I ran:
apt-get update
apt-get install -y gcc-11 g++-11
export CC=gcc-11
export CXX=g++-11Resolved fatal error: crypt.h: No such file or directory/ninja: build stopped: subcommand failed as is detailed in ColBERT issue #371.
Functions Selected for Profiling
I determined which functions to profile by trial and error, adding/removing the @profile decorator to see which function was being called. Again, lots of Claude assistance was needed. Here are the filenames and method names that I chose to profile:
ColBERT
| Filename | Method |
|---|---|
| indexer.py | index |
| indexer.py | __launch |
| indexing/collection_indexer.py | encode |
| indexing/collection_indexer.py | run |
| indexing/collection_indexer.py | setup |
| indexing/collection_indexer.py | __sample_pids |
| indexing/collection_indexer.py | __sample_embeddings |
| indexing/collection_indexer.py | encoder.encode_passages |
| infra/launcher.py | launch |
| infra/launcher.py | launch_without_fork |
| infra/launcher.py | run_process_without_mp |
| infra/launcher.py | callee |
RAGatouille
| Filename | Method |
|---|---|
| RAGPretrainedModel.py | _process_corpus |
| RAGPretrainedModel.py | model.index |
| models/colbert.py | ModelIndexFactory.construct |
| models/index.py | PLAIDModelIndex.__init__ |
| models/index.py | PLAIDModelIndex.construct |
| models/index.py | PLAIDModelIndex.build |
| models/index.py | PLAIDModelIndex.indexer.index |
Note that in RAGatouille, PLAIDModelIndex.indexer is of class Indexer which is imported from ColBERT, so I understood this to be the “bridge” between the RAGatouille and ColBERT repos during profiling.
Scripts
Here’s the script for indexing using ColBERT:
import colbert
from colbert import Indexer, Searcher
from colbert.infra import Run, RunConfig, ColBERTConfig
from colbert.data import Queries, Collection
from datasets import load_dataset
from memory_profiler import profile
@profile
def _index(indexer, name, collection):
return indexer.index(name=name, collection=collection, overwrite=True)
def main():
nbits = 2
ndocs = 100_000
dataset_name = "Genomics"
index_name = f'{dataset_name}.{nbits}bits'
passages = load_dataset("UKPLab/dapr", f"{dataset_name}-corpus", split="test")
checkpoint = 'answerdotai/answerai-colbert-small-v1'
with Run().context(RunConfig(nranks=1, experiment='notebook')):
config = ColBERTConfig(doc_maxlen=256, nbits=nbits, kmeans_niters=4, avoid_fork_if_possible=True)
indexer = Indexer(checkpoint=checkpoint, config=config)
_index(indexer, index_name, passages[:ndocs]["text"])
if __name__ == '__main__':
main()and the script for RAGatouille:
from memory_profiler import profile
from datasets import load_dataset
from ragatouille import RAGPretrainedModel
dataset_name = "Genomics"
passages = load_dataset("UKPLab/dapr", f"{dataset_name}-corpus", split="test")
RAG = RAGPretrainedModel.from_pretrained("answerdotai/answerai-colbert-small-v1")
ndocs=250_000
@profile
def _index():
return RAG.index(
index_name=f"{dataset_name}_index",
collection=passages[:ndocs]["text"],
document_ids=passages[:ndocs]["_id"],
use_faiss=True # or False
)
_index()Finally, here’s the terminal command to run the scripts and profile them:
python -m memory_profiler ../colbert_index_2M.py > ../colbert_2M_RTX6000Ada.txtProfiling Results
The profile logs were 400+ lines each (you can see the full files here) so I have only included some of the lines with non-zero memory changes. I have showed the starting memory, memory increment and final memory.
Here’s how I’m interpreting the profiler logs–given this log:
Filename: /home/RAGatouille/ragatouille/models/index.py
Line # Mem usage Increment Occurrences Line Contents
=============================================================
198 3406.4 MiB 3406.4 MiB 1 @profile
199 def _index_with_profiling(indexer, name, collection, overwrite):
200 4872.2 MiB 1465.8 MiB 1 return indexer.index(name=name, collection=collection, overwrite=overwrite)I would interpret that to mean that before indexer.index was called, 3406.4 MB memory was used, and the indexer.index call increased it by 1465.8 MB to 4872.2 MB.
colbert/indexer.py/indexer.index
For RAGatouille, this call takes place in ragatouille/models/index.py.
It’s interesting to note that even before indexer.index is called, the starting memory varies between raw ColBERT and RAGatouille. Most notably, for 2M documents, ColBERT starts at ~4GB while RAGatouille starts at ~8 GB.
Even more interesting, the memory increments for ColBERT are 2x to 35x smaller than RAGatouille for each collection size.
| Indexing Method | Document Size | Starting Memory | Memory Increment | Final Memory |
|---|---|---|---|---|
| ColBERT | 100k | 1596.9 MB | 36.7 MB | 1633.6 MB |
| ColBERT | 250k | 1754.0 MB | 92.8 MB | 1846.8 MB |
| ColBERT | 500k | 2072.1 MB | 199.1 MB | 2271.2 MB |
| ColBERT | 1M | 2707.3 MB | 421.9 MB | 3129.2 MB |
| ColBERT | 2M | 4000.6 MB | 876.4 MB | 4877.1 MB |
RAGatouille (use_faiss=True) |
100k | 2114.2 MB | 1320.1 MB | 3434.3 MB |
RAGatouille (True) |
250k | 2592.5 MB | 1175.0 MB | 3767.5 MB |
RAGatouille (True) |
500k | 3405.0 MB | 1430.0 MB | 4835.0 MB |
RAGatouille (use_faiss=False) |
100k | 1750.9 MB | 1203.9 MB | 2954.8 MB |
RAGatouille (False) |
250k | 2597.4 MB | 1341.4 MB | 3938.8 MB |
RAGatouille (False) |
500k | 3406.4 MB | 1465.8 MB | 4872.2 MB |
RAGatouille (False) |
1M | 5040.1 MB | 1593.3 MB | 6633.3 MB |
RAGatouille (False) |
2M | 8354.7 MB | 1882.0 MB | 10236.8 MB |
colbert/indexing/collection_indexer.py/encoder.encode_passages
encoder.encode_passages involves the following code:
def encode_passages(self, passages):
Run().print(f"#> Encoding {len(passages)} passages..")
if len(passages) == 0:
return None, None
with torch.inference_mode():
embs, doclens = [], []
for passages_batch in batch(passages, self.config.index_bsize * 50):
embs_, doclens_ = self.checkpoint.docFromText(
passages_batch,
bsize=self.config.index_bsize,
keep_dims="flatten",
showprogress=(not self.use_gpu),
pool_factor=self.config.pool_factor,
clustering_mode=self.config.clustering_mode,
protected_tokens=self.config.protected_tokens,
)
embs.append(embs_)
doclens.extend(doclens_)
embs = torch.cat(embs)
return embs, doclensIIUC, this is calling docFromText on the ColBERT model (answerai-colbert-small-v1 in our case). I would expect raw ColBERT and RAGatouille to experience equal memory change during this method call but RAGatouille uses 10-15% more memory for each dataset size.
| Indexing Method | Document Size | Initial Memory | Memory Change | Final Memory |
|---|---|---|---|---|
| ColBERT | 100k | 732.9 MB | 1502.4 MB | 2235.3 MB |
| ColBERT | 250k | 829.7 MB | 1991.1 MB | 2820.8 MB |
| ColBERT | 500k | 1000.2 MB | 2549.8 MB | 3550.0 MB |
| ColBERT | 1M | 1351.6 MB | 3462.0 MB | 4813.6 MB |
| ColBERT | 2M | 1997.3 MB | 4692.3 MB | 6689.6 MB |
RAGatouille (use_faiss=True) |
100k | 2115.0 MB | 1677.3 MB | 3792.3 MB |
RAGatouille (True) |
250k | 2593.5 MB | 2279.7 MB | 4873.2 MB |
RAGatouille (True) |
500k | 3405.1 MB | 3004.6 MB | 6409.6 MB |
RAGatouille (use_faiss=False) |
100k | 1751.0 MB | 1685.6 MB | 3436.6 MB |
RAGatouille (False) |
250k | 2597.9 MB | 2270.4 MB | 4868.3 MB |
RAGatouille (False) |
500k | 3406.4 MB | 3003.8 MB | 6410.2 MB |
RAGatouille (False) |
1M | 5040.7 MB | 3915.3 MB | 8956.0 MB |
RAGatouille (False) |
2M | 8355.1 MB | 5349.5 MB | 13704.6 MB |
colbert/indexing/collection_indexer.py/_sample_embeddings
encode_passages is called from inside _sample_embeddings. For ColBERT, _sample_embeddings has different starting/final memory values than _encode_passages while for RAGatouille they are the same.
For example, for 100k documents using raw ColBERT, _sample_embeddings increases memory by 797 MB while for encoder.encode_passages the memory increases by 1488.8MB.
For 100k using RAGatouille, both memory increases the same (1677.3 MB for use_faiss=True and 1685.6 MB for use_faiss=False). I’m not sure what this means so I asked Claude and got the response:
This discrepancy reveals memory reuse patterns between function calls. In ColBERT, the 1488.8 MB used by
encode_passagesis partially freed before returning to_sample_embeddings, resulting in a net increase of 797 MB. In RAGatouille, the memory appears to be retained between calls, showing the same 1677.3 MB increase at both levels.
| Indexing Method | Document Size | Initial Memory | Memory Change | Final Memory |
|---|---|---|---|---|
| ColBERT | 100k | 732.9 MB | 813.8 MB | 1546.7 MB |
| ColBERT | 250k | 829.7 MB | 809.0 MB | 1638.7 MB |
| ColBERT | 500k | 1000.2 MB | 770.1 MB | 1770.3 MB |
| ColBERT | 1M | 1351.6 MB | 813.3 MB | 2164.9 MB |
| ColBERT | 2M | 1997.3 MB | 782.4 MB | 2779.7 MB |
RAGatouille (use_faiss=True) |
100k | 2115.0 MB | 1677.3 MB | 3792.3 MB |
RAGatouille (True) |
250k | 2593.5 MB | 2279.7 MB | 4873.2 MB |
RAGatouille (True) |
500k | 3405.1 MB | 3004.6 MB | 6409.6 MB |
RAGatouille (use_faiss=False) |
100k | 1751.0 MB | 1685.6 MB | 3436.6 MB |
RAGatouille (False) |
250k | 2597.9 MB | 2270.4 MB | 4868.3 MB |
RAGatouille (False) |
500k | 3406.4 MB | 3003.8 MB | 6410.2 MB |
RAGatouille (False) |
1M | 5040.7 MB | 3915.3 MB | 8956.0 MB |
RAGatouille (False) |
2M | 8355.1 MB | 5349.5 MB | 13704.6 MB |
colbert/indexing/collection_indexer.py/setup
A similar pattern for setup, within which _sample_embeddings is called. Raw ColBERT seems more efficient in releasing memory while RAGatouille retains it.
| Indexing Method | Document Size | Initial Memory | Memory Change | Final Memory |
|---|---|---|---|---|
| ColBERT | 100k | 727.9 MB | 817.5 MB | 1545.5 MB |
| ColBERT | 250k | 815.7 MB | 816.4 MB | 1632.1 MB |
| ColBERT | 500k | 978.2 MB | 787.9 MB | 1766.1 MB |
| ColBERT | 1M | 1305.6 MB | 840.2 MB | 2145.8 MB |
| ColBERT | 2M | 1966.3 MB | 822.2 MB | 2788.5 MB |
RAGatouille (use_faiss=True) |
100k | 3434.3 MB | 1677.3 MB | 3792.3 MB |
RAGatouille (True) |
250k | 3767.5 MB | 2279.7 MB | 4873.2 MB |
RAGatouille (True) |
500k | 4835.0 MB | 3004.6 MB | 6409.6 MB |
RAGatouille (use_faiss=False) |
100k | 2954.8 MB | 1685.6 MB | 3436.6 MB |
RAGatouille (False) |
250k | 3938.8 MB | 2270.4 MB | 4868.3 MB |
RAGatouille (False) |
500k | 4872.2 MB | 3003.8 MB | 6410.2 MB |
RAGatouille (False) |
1M | 6633.3 MB | 3915.3 MB | 8956.0 MB |
RAGatouille (False) |
2M | 10236.8 MB | 5349.5 MB | 13704.6 MB |
colbert/indexing/collection_indexer.py/train
IIUC, this function call finds centroids based on a sample of document token embeddings. Interesting to note that the memory change for raw ColBERT is smallest for 1M documents (87.2 MB) and for RAGatouille, 2M docs is the smallest (23.4 MB). For most collection sizes, RAGatouille uses 40-50% more memory for this operation.
| Indexing Method | Document Size | Initial Memory | Memory Change | Final Memory |
|---|---|---|---|---|
| ColBERT | 100k | 1545.5 MB | 115.8 MB | 1661.3 MB |
| ColBERT | 250k | 1632.1 MB | 128.8 MB | 1760.9 MB |
| ColBERT | 500k | 1766.1 MB | 124.3 MB | 1890.4 MB |
| ColBERT | 1M | 2145.8 MB | 87.2 MB | 2233.0 MB |
| ColBERT | 2M | 2788.5 MB | 133.5 MB | 2921.9 MB |
RAGatouille (use_faiss=True) |
100k | 3792.3 MB | 179.6 MB | 3971.9 MB |
RAGatouille (True) |
250k | 4873.2 MB | 182.7 MB | 5055.9 MB |
RAGatouille (True) |
500k | 6409.6 MB | 174.1 MB | 6583.8 MB |
RAGatouille (use_faiss=False) |
100k | 3436.6 MB | 175.9 MB | 3612.6 MB |
RAGatouille (False) |
250k | 4868.3 MB | 181.5 MB | 5049.8 MB |
RAGatouille (False) |
500k | 6410.2 MB | 179.2 MB | 6589.4 MB |
RAGatouille (False) |
1M | 8956.0 MB | 191.5 MB | 9147.5 MB |
RAGatouille (False) |
2M | 13704.6 MB | 23.4 MB | 13728.1 MB |
colbert/indexing/collection_indexer.py/index
This is one of the more interesting results—raw ColBERT has a positive memory change during this operation (which IIUC is the indexing of all document token embeddings) while all RAGatouille index() operations actually reduce the memory usage. Not sure what that means. The final memory for raw ColBERT is less than RAGatouille.
| Indexing Method | Document Size | Initial Memory | Memory Change | Final Memory |
|---|---|---|---|---|
| ColBERT | 100k | 1661.3 MB | 287.0 MB | 1948.3 MB |
| ColBERT | 250k | 1760.9 MB | 263.5 MB | 2024.4 MB |
| ColBERT | 500k | 1890.4 MB | 371.9 MB | 2262.2 MB |
| ColBERT | 1M | 2233.0 MB | 599.9 MB | 2832.9 MB |
| ColBERT | 2M | 2921.9 MB | 958.0 MB | 3880.0 MB |
RAGatouille (use_faiss=True) |
100k | 3971.9 MB | -536.3 MB | 3435.6 MB |
RAGatouille (True) |
250k | 5055.9 MB | -1375.8 MB | 3680.1 MB |
RAGatouille (True) |
500k | 6583.8 MB | -1936.3 MB | 4647.5 MB |
RAGatouille (use_faiss=False) |
100k | 3612.6 MB | -652.4 MB | 2960.2 MB |
RAGatouille (False) |
250k | 5049.8 MB | -1112.5 MB | 3937.3 MB |
RAGatouille (False) |
500k | 6589.4 MB | -1906.8 MB | 4682.6 MB |
RAGatouille (False) |
1M | 9147.5 MB | -2917.3 MB | 6230.1 MB |
RAGatouille (False) |
2M | 13728.1 MB | -4910.2 MB | 8817.9 MB |
colbert/indexing/collection_indexer.py/finalize
This function maps passage IDs to centroid IDs—one of the efficiencies of the PLAID indexing approach. Within each approach (raw ColBERT and RAGatouille) the memory change varies drastically between less than 0 and up to ~500MB.
| Indexing Method | Document Size | Initial Memory | Memory Change | Final Memory |
|---|---|---|---|---|
| ColBERT | 100k | 1948.3 MB | 35.1 MB | 1983.3 MB |
| ColBERT | 250k | 2024.4 MB | -0.4 MB | 2024.0 MB |
| ColBERT | 500k | 2262.2 MB | 59.2 MB | 2321.5 MB |
| ColBERT | 1M | 2832.9 MB | 201.5 MB | 3034.4 MB |
| ColBERT | 2M | 3880.0 MB | 490.2 MB | 4370.2 MB |
RAGatouille (use_faiss=True) |
100k | 3435.6 MB | -1.3 MB | 3434.3 MB |
RAGatouille (True) |
250k | 3680.1 MB | 87.4 MB | 3767.5 MB |
RAGatouille (True) |
500k | 4647.5 MB | 187.5 MB | 4835.0 MB |
RAGatouille (use_faiss=False) |
100k | 2960.2 MB | -5.3 MB | 2954.8 MB |
RAGatouille (False) |
250k | 3937.3 MB | 1.5 MB | 3938.8 MB |
RAGatouille (False) |
500k | 4682.6 MB | 189.6 MB | 4872.2 MB |
RAGatouille (False) |
1M | 6230.1 MB | 403.2 MB | 6633.3 MB |
RAGatouille (False) |
2M | 8817.9 MB | 1418.9 MB | 10236.8 MB |
Indexing Time
I didn’t measure runtime for each run, but some observations:
- During passage encoding (25k passages per iteration) ColBERT took about 20 seconds/it and RAGatouille took about 110 seconds/it. Note that without profiling ColBERT took about 9/seconds/it and RAGatouille 12 seconds/it.
- ColBERT encoding lasted 4, 10, 20, 40 and 80 iterations for 100k, 250k, 500k, 1M and 2M docs. RAGatouille always overshot it (e.g. 14 iters for 250k docs or 22 iters for 500k docs).
- Overall ColBERT profiling took ~2 hours while RAGatouille took ~16 hours.
- It took a lot of time before the final encoding takes place, I think that’s because of the initial “planning” step that ColBERT and RAGatouille both do.
Indexing 10k Documents (PyTorch vs FAISS K-means Clustering)
While I was experimenting indexing scripts with 10k documents I noticed curious behavior. For 10k documents, with use_faiss=False, RAGatouille attempts to use PyTorch for K-means clustering. The memory usage for encoder.encode_passages during this attempt:
Line # Mem usage Increment Occurrences Line Contents
=============================================================
146 1849.2 MiB 1849.2 MiB 1 @profile
147 def _encode_passages_profiled(*args, **kwargs):
148 2675.7 MiB 826.5 MiB 1 return self.encoder.encode_passages(*args, **kwargs)It then runs into an OOM error:
PyTorch-based indexing did not succeed with error: CUDA out of memory. Tried to allocate 27.55 GiB. GPU 0 has a total capacity of 47.51 GiB of which 4.88 GiB is free.And switches to FAISS K-means. The memory usage for encoder.encode_passages changes (note the drop from an increase of 826.5 MB to an increase of 373 MB, but an increase in initial memory usage from 1849.2 MB to 2652.6MB):
Line # Mem usage Increment Occurrences Line Contents
=============================================================
146 2652.6 MiB 2652.6 MiB 1 @profile
147 def _encode_passages_profiled(*args, **kwargs):
148 3025.6 MiB 373.0 MiB 1 return self.encoder.encode_passages(*args, **kwargs)When I run the script with use_faiss=True, the encoder.encode_passages memory usage reflects the PyTorch attempt, whereas I would expect the memory increase to be 373 MB:
Line # Mem usage Increment Occurrences Line Contents
=============================================================
146 1853.4 MiB 1853.4 MiB 1 @profile
147 def _encode_passages_profiled(*args, **kwargs):
148 2678.8 MiB 825.4 MiB 1 return self.encoder.encode_passages(*args, **kwargs)Final Thoughts
This exercise has left me with more questions than answers that I need to explore:
- Is this the best way to go about profiling memory?
- Am I interpreting the memory profiling results correctly?
- Why does RAGatouille have a higher initial memory before indexing starts?
- Why does RAGatouille retain more memory after indexing than ColBERT?
- Why does RAGatouille memory usage drastically decrease during
index()? - Why does RAGatouille max out CUDA memory for 10k documents? Related to Issue #247.
- Why does RAGatouille’s memory usage when
use_faiss=Truematch PyTorch K-means’ memory usage and not the FAISS K-means’ memory usage after PyTorch’s attempt fails with OOM?
Additionally, and probably relatedly, I still haven’t figured out what is causing the large memory spike in the diagram below:
The largest memory value profiled while indexing 250k docs using RAGatouille was 5 GB but the chart shows a spike up to ~8GB. Where’s the ghost 3GB?
TBD.